2nd semester
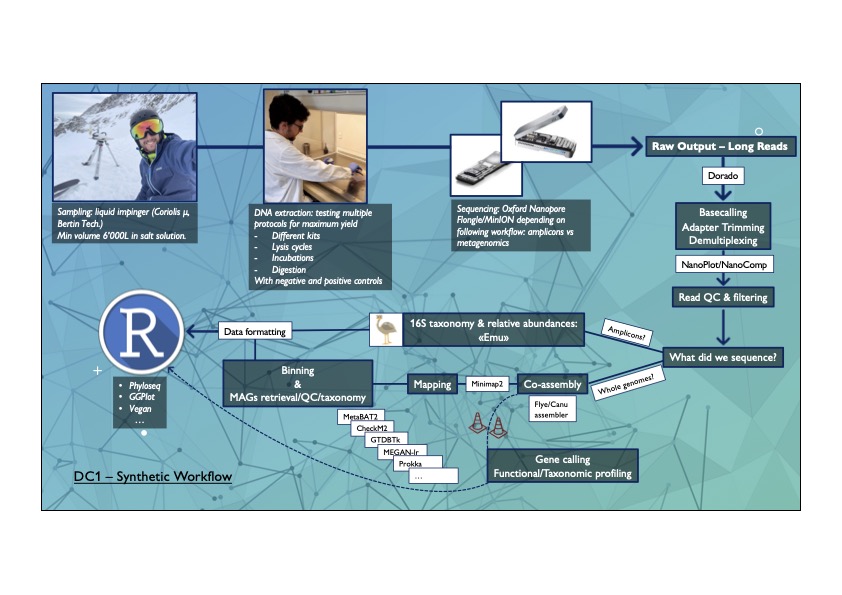
DC1 ALESSANDRO CUZZERI OCTOBER 23 - MARCH 24
Month 7: (October 2023) – Extensive research in the field of bioinformatics, related to the outcome of the crash course attended at the end of September 2023, particularly in the direction of HPC (High Performance Computing) metagenomic tools implementation and compatibility.
Month 8: (November 2023) – Obtained access to the University of Innsbruck’s HPC resources. Started testing different bioinformatic tools and pipelines, in light of the newly gained computational resources, to optimize processing speed and especially output quality. As anticipated in the previous report, preliminary data on cryoconite communities time series is available for processing and analyses. Started a biostatistics course ecology-oriented to gain deeper methodological knowledge to be implemented in the future paper. Collected air samples from the Stubai Glacier to test extraction efficiency.
Month 9: (December 2023) – Preliminary data briefly presented during midterm check. First extractions on test samples reveal very low DNA contents. Continuous research on bioinformatics and biostatistics.
Month 10: (January 2024) – More DNA extractions, various protocol modifications to maximize DNA retrieval. Continuous research on bioinformatics and biostatistics. Different assembly and binning testing.
Month 11: (February 2024) – High school outreach activity in Novara, Italy. Second ICEBIO meeting in Grenoble, France. Continuous research on bioinformatics and biostatistics.
Month 12: (March 2024) – Reshaping of research question #1 hypotheses in light of new inputs during the Grenoble meeting and consistently low DNA in air samples.

DC6 KLARA KOHLER NOVEMBER 23 - APRIL 24
Month 7 (November):
In November, I attended the International Glaciology Society Nordic Branch conference in Finland, which was held at Aalto University. At the conference I presented a poster about the IceBio network and the research ideas for my project. In the lab, I finalized the methods for (inorganic) P and Si analysis with the Gallery Analyzer.
Month 8:
In December, I continued to test the methods for my nutrient analysis and completed the methods for measuring inorganic N and Fe species with the Gallery Analyzer. I also tested methods for measuring the total P, N, Si and Fe concentration in a sample, from which I can later calculate the organic nutrient content.
Month 9:
In January, I started testing my methods on the first sediment sample from Greenland, which was collected in 2022. I measured the nutrient content (inorganic P, Si, N and Fe concentrations) of the water above the sediment in the sample jar and the pore water that I extracted from the sediment using Rhizon samplers. In the middle of the months I participated in the first part of the PhD course "Scientific Teaching" at Aarhus University in Aarhus. At the end of the months, I attended the annual science meeting of the Deep Purple project, where I presented my project and received valuable input from colleagues working with similar research topics and methods.
Month 10:
In February, I continued the work with the Greenland sample and measured the total P, Si, N and Fe concentration of the overlaying and pore water. I also worked on the second part of the PhD course "Scientific Teaching". At the end of the month, I went to Grenoble for the second IceBio network event, where we learned about omics and about sampling and experimental design.
Month 11:
In March, I tested extraction methods on the Greenland sample to measure the concentrations of P, Si, N and Fe species bound in the sediment. I spent the last two weeks of the month at the GFZ in Potsdam to learn different mineralogical techniques that will be useful for further analysis of my sediment and rock samples.
Month 12:
In April, I continued to work on finalizing the extraction methods. I also spent a lot of time this month preparing for the field work. We had several organizational meetings, and I had to order, wash and pack the equipment I would need in the field. I also took part in riffle training and a first aid course, both of which are required for fieldwork in Greenland.

DC7 MARCO AJMAR DEC 23 - JUNE 24
Month 7 (Dec 15 - Jan 15):
IICP-MS data analyses, literature reading about glacier rivers nutrient cycles and microbiology.
Month 8: (Jan 15-Feb. 15): Tested different particles filtration protocols and attended the annual Deep Purple Project meeting in Portugal. Collaborated with Prof. M. A. Diaz (Ohio State University) to analyse proglacial lake sediments samples from Greenland using FT-IR spectroscopy, XRD and SEM at the GFZ.
Month 9 (15th February – 15th March): Prepared and carried out an outreach activity in an Italian highschool (together with DC1 Alessandro Cuzzeri). Attended the 2nd IceBio meeting in Grenoble about experimental design, omics and data handling. Analysed ice and snow samples (previously collected by the GFZ team in Svalbard during winter 2023) for dissolved inorganic and organic P at Aarhus University.
Month 10 (15th March – 15th April): Attended a two-week course at the GFZ focused on characterization methods for mineralogy (optical microscopy, XRD and TEM) and particles chemical mapping (SEM). Ordered a dissolved silica sensor from IceBio partner ClearWater Sensors and a multiparameter sensor for pH, turbidity, conductivity and dissolved oxygen monitoring.
Month 11 (15th April – 15th May): Packed for Summer fieldwork, attended two safety courses (Polar bear protection at AWI and 1st aid at the GFZ) and transported sampling gear to Aarhus University.
Month 12 (15th May – 15th June): Planned fieldwork in Iceland for August 2024 and prepared a poster presentation for the PhD days at the GFZ. Brief introduction on ICP-OES and pH/turbidity sensors.

DC5 ANIRBAN MAJUMDER JAN 24 - JUNE 24
Month 7 (January 24):
At the beginning of the new year, I planned the metabolite extraction lab work for the future. Simultaneously I was also working on my literature review.
Month 8 (Feb 2024): At the start of this month, I began the extraction of metabolites from my samples. Then I attended the Icebio network event for bioinformatics training at Grenoble, France.
Month 9 (Mar 2024): I continued my metabolite extraction and took the samples to analyse in mass spectrometry with our collaborator in BAM. Later, I started the preparation for my upcoming Greenland summer fieldwork. In the mid of March, I attended a workshop at the University of Cologne for bioinformatics metabolomics analysis.
Month 10 (Apr 2024): I continued packing for the Greenland fieldwork. At the end of this month, I got first-aid and polar bear safety training.
Month 11 (May 2024): At the beginning of May, I planned and helped with the final packing and shipment of the lab and camp materials for Greenland fieldwork. 1 went to our collaborator BAM to analyse the metabolite samples in mass spectrometry in mid-May. Later, I started writing my transfer report for the first year.
Month 12 (June 2024): I completed the first draft of my transfer report including my literature review. In mid-June, I also presented a poster on PhD day at GFZ.

DC8 SILJE WAALER: Feb 24 - July 24
Month 7: - February was used to finish sampling plan for the field season in Kongsfjorden 2024. This also included buying home consumables and equipment needed. Because we are looking at trace metals and mercury, thorough rinsing procedures had to be performed on consumables. Protocols for different aspects of the sampling was also made including DNA and RNA sampling. Moreover, a presentation of my research plan was made and presented at the annual METALLICA meeting at Fram Senteret, Tromsø.
Month 8 March was used for the continues work on rinsing of consumables. Moreover, the first load of equipment was packed and sent to Svalbard to be ready for the pre-melt campaign in Kongsfjorden. Nutrient samples generated through the pilot study on the dissolution experiment with glacial flour was analysed using the POLAR MAGIC lab’s new discrete and segmented flow analysers from SEAL.
Month 9 April was used for data analysis of the data obtained through the pilot study on the dissolution experiment and finishing off most of the prepping of consumables for the remaining field campaigns in Kongsfjorden 2024. In late April, the pre-melt campaign was started in Kongsfjorden targeting both water chemistry and microbial community composition and activity through out the fjord.
Month 10 May was used for finishing the pre-melt campaign in Kongsfjorden successfully retrieving DNA and RNA samples from under the sea ice in front of Kronebreen. Rest of May was used for preparing for the upcoming field campaigns in Kongsfjorden including more rinsing of consumables related to trace metals and mercury sampling. Moreover, the last shipment of equipment was sent off to Svalbard during this period.
Month 11 June was used to finish off the last round of prepping before setting off for the early melt campaign in Kongsfjorden. During the field campaign a whole fjord transect of Kongsfjorden was performed incl. water chemistry and microbiology in the water column along with sediment core sampling both porewater and bulk concentrations of trace metals, mercury and microbial community composition. Lastly suspended sediment was collected from three major glacial meltwater outflows in Kongsfjorden. The fjord transect was performed in collaboration with Norwegian Polar Institute and the MSCA SEASOL project
Month 12 July was used for finishing the last part of the early melt campaign and preparing for the peak melt campaign in Kongsfjorden. Late July the second fjord transect was started reproducing the sampling procedure of the first fjord transect exluding sediment core sampling.

DC4 LARS VAN DIJK: Feb 2024 - July 2024
Month 6 – February 2024: In the sixth month (February), I worked on the PhD course "Scientific Teaching" at Aarhus University, which ran until May, and participated in the second ICEBIO network event on “Experimental Design and Data handling” and “The World of Omics in the Cryosphere”.
Month 7 – March 2024: In the seventh month, microbial isolates from a pilot experiment were sent for species identification via Sanger sequencing. I also got introduced to working in a radioactive isotope laboratory, attended a workshop on “Managing Your PhD Project and Handling Stress”, and presented my progress and project at our Wednesday seminar at work.
Month 8 – April 2024: In the eighth month (April), I performed DNA extractions of cryoconite material and prepared the samples for 16S amplicon sequencing. Additionally, I began working with DC1 and DC11 on one of the first ICEBIO deliverables: "Workflow for PCR-Amplicon, Metagenomic, and Total RNA Analyses of Glacier Samples," which we finalized at the beginning of the following month.
Month 9 – May 2024: In the ninth month (May), I received the sequencing data from all my samples, processed the data using the established workflow from the ICEBIO deliverable, and performed initial analyses to gain insights into my results. Also, I completed the PhD course "Scientific Teaching" and, at the end of the month, visited my former high school for an outreach event where I discussed my journey towards pursuing a PhD, my PhD project, and ICEBIO.
Month 10 – June 2024: In the tenth month (June), I began collecting, organizing, and reviewing scientific literature for the next deliverable I am responsible for: “Catalogue of the Active Microbial Diversity on Glacier Surfaces”. Furthermore, together with the supervisors, DC1 and I prepared a collaborative project on pesticidal degradation by microbial communities in cryoconite on the Forni Glacier, with fieldwork scheduled for the end of August.
Month 11 – July 2024: In the eleventh month, some of the (slow-growing) microbial isolates obtained in the fourth month have been purified and are now ready for species identification, other isolates will be done in the next month - but first, it is time to take a break and enjoy some annual leave.

DC3 ANNIKA MORISCHE: Feb 2024 - July 2024
Month 7 (February 2024) I started the beginning of the second semester off by fine-tuning the quality control measures for analysing my endometabolome samples. This involved perfecting protocols to accurately reflect the metabolic changes in glacial microbiological communities facing various environmental stressors. My focus was on optimizing how to best collect, store, and process samples to ensure the metabolites stay intact.
Month 8 (March 2024) This month was all about detailing fieldwork plans for the upcoming expedition to the Greenland Ice Sheet including arranging the order of equipment needed for sample collection and processing. In addition, The Deep Purple network event was a goldmine of ideas, where discussions with experts in organic chemistry and glacier-related omics sparked valuable insights. I started mapping out how to link my future metabolomic findings with genomic data, setting the stage for a workflow that integrates both metabolic and genetic data.
Month 9 (April 2024) In month nine, I focused on finalizing the logistics for fieldwork, ensuring all necessary equipment and lab ware were ordered and prepared. We also secured the infrastructure needed for the safe transport and storage of samples. Additionally, I participated in critical fieldwork safety training, including a course on firearm use and Arctic first aid.
Month 10 (May 2024) As our Greenland expedition approached, packing and organizing gear for the team heading to the ice shield was most important. I got the chance to gain deeper insights into coordinating projects with other fieldwork participants.
Month 11 (June 2024) I conducted a test study in preparation for the 2024 field season, working with microalgae cultures of the genus Ancylonema and Deuterostichococcus. By labelling algal cells with ¹³C - sodium bicarbonate we strive to unveil novel insight about microbial carbon flux from glaciers and secondary metabolite production. The tracing of carbon through cellular processes reveals active metabolic pathways in glacial primary producers and advances our understanding of fundamental algal metabolism. The study also involved identifying secondary metabolites, including potential antibiotics, to explore the controls on their biosynthesis and environmental effects. The data obtained captured a diverse array of endometabolites and offered valuable insights into the chemical diversity of glacier ecosystems. Additionally, I tested different bicarbonate concentrations to ensure sufficient levels of labelled metabolites to be detected during analysis. For example, the processed data indicated the successful incorporation of ¹³C into cell wall compounds.
Month 12 (July 2024) Fieldwork in Kangerlussuaq started strong with nearly daily trips to the Greenlandic ice shield and the surrounding area. For my incubation experiments, I gathered surface ice and cryoconite material. As an example of field sample preparation, ice samples were first melted and homogenized, then divided into triplicate 200 mL (for endometabolomics) and 1000 mL (for exometabolomics) gas-tight glass bottles. The samples were subsequently spiked with ¹³C-sodium bicarbonate and incubated directly on the ice. Control samples were prepared without bicarbonate at the start of the experiment, while ¹²C-sodium bicarbonate incubations were conducted and sampled alongside their ¹³C counterparts, serving as reference points for ¹³C incorporation into metabolites. After a duration of 24 and 48 hours, I quickly filtered the samples and quenched the metabolic activity, wrapping up a successful and hands-on three-weeks in the field. After extracting and preparing my samples for metabolomic and lipidomic analysis via mass spectrometry, the generated data promises to establish a comprehensive database of compounds produced by surface-ice primary producers.
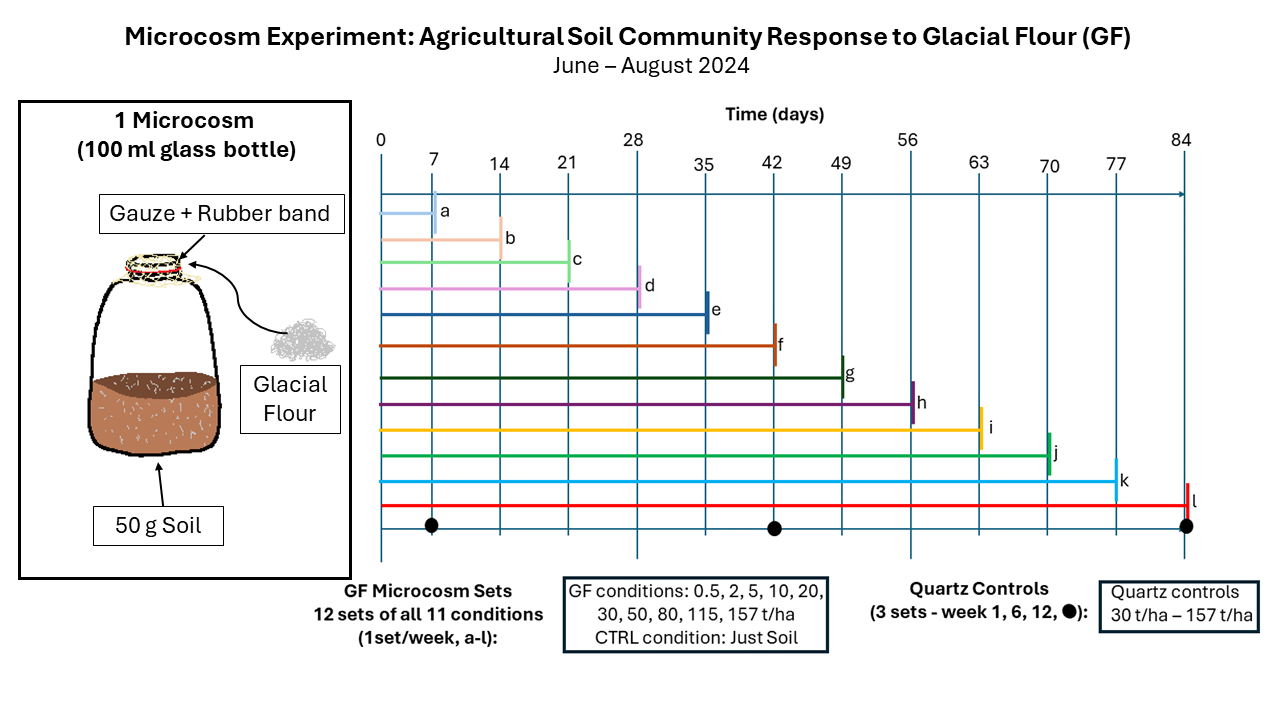
DC9 KARA SAMPSELL: March 2024 - August 2024
Month 7 (March 2024) I finalized a plan for a microcosm experiment designed to investigate the response of microbial communities in agricultural soil to a range of ten glacial flour quantities over a period of twelve weeks. During this month, I collected and prepared the materials to be used in this experiment such as the agricultural soil, 100 ml glass bottles (acid-rinsed and calcinated), and quartz to be used a control amendment.
Month 8 (April 2024) I continued preparing for the microcosm experiment by completing initial analyses on the water content and holding capacity of the soil. I arranged for the control quartz material to be finely ground by a collaborator to mimic the glacial flour grain size. It was also in this month that I prepared my initial glacial flour samples for metagenomics sequencing (Illumina). The run was successful, and I received the data at the end of the month.
Month 9 (May 2024) I worked preparing glacial flour DNA extracts for 16S amplicon sequencing but encountered problems that stemmed from poor amplification and required further troubleshooting. I gave an outreach presentation to students at the American School of Grenoble in the middle of the month and did final preparations to start the microcosm experiment at the beginning of June.
Month 10 (June 2024) On June 4th I set up the soil and microcosm experiment with the help of my supervisor. It will conclude at the end of August 2024. In each week of the experiment, I replace moisture lost from the microcosms, perform destructive sampling, and record the temperature of the room. I began extracting DNA from these samples. I completed my third French course, an online course on Research Ethics, and an Introduction to AI course during this month. I completed 16S qPCR on the glacial flour DNA extracts. I began working on the downstream analysis of my metagenomics data and obtained some preliminary results. I presented my project quickly to the other doctoral students in our faculty during the annual PhD Day.
Month 11 (July 2024) IOn July 9th I attended my yearly supervisory meeting required by the doctoral school (CSI meeting) with a committee comprised by my supervisor and other experts/non-experts in the field. I prepared a report and presentation for their review and received helpful commentary on the direction of my research. During this month I also optimized and performed 16S qPCR on the first five weeks of DNA-extracts from my microcosm experiment to gain early insight on potential effects to the microbial community over time. Regular maintenance of the microcosms continued.
Month 12 (August 2024) I analysed the 16S qPCR results and continued DNA extractions from the microcosm samples. Based on discussion in my CSI meeting, I met with an expert in our lab to plan for gas measurements from one set of my microcosms in the Fall. I worked on preparing 16S amplicon sequencing again for the glacial flours and the first few weeks of the microcosm experiment. I continued researching tools and implementation for the metagenomics data analyses and maintaining the microcosm experiment.

DC2 HARPREET SINGH: March 2024 - AUGUST 2024
Months 7, 8 and 9 (March - May 2024) The first 3 months were spent in designing the research objectives and hypotheses. The extensive literature survey led to finalizing 4 objectives. All the information supportive of the objectives and the required work plan was compiled. Simultaneously other training programs were attended to gain competencies in artificial intelligence and data analyses.
Month 10 (June 2024) We prepared for fieldwork in Ny-Alesund, Svalbard where we dug the snow pits and collected the snow samples from Midtre Lovenbreen. The samples were filtered in the research station and stored at -20. We also collected some samples for the microcosm studies (around 40kg snow).
Months 11 and 12 (July - August 2024) Upon returning from Svalbard, we started processing our samples to extract the DNA and RNA for sequencing. Around 105 samples were processed and stored at -20. All the DNA samples were subjected to library preparation for the metagenome sequencing. We used our inhouse, Illumina NextSeq1000 sequencer to run our samples. We carried out 2 runs of sequencing with 50-53 samples per flow cell. Both the runs were successfully conducted and we have stored the data in our server. Now we have to proceed with the quality check and analyses of our data.

DC2 NASIM KASHIRI: MAY 2024 - OCToBER 2024
Month 7 (May): I completed an internship at the main branch of our company in Hamburg, where I gained hands-on experience with MIC (Minimum Inhibitory Concentration) testing and other essential techniques related to antibiotic resistance analysis. This experience also allowed me to become familiar with the operational environment at the Hamburg branch. Afterward, I began the first part of my secondment at Aarhus University.
Month 8 (June): I spent the month at Aarhus University, where I conducted DNA extraction and performed whole genome sequencing using nanopore technology for the first time. Following this, I began learning gene mining techniques to analyze the sequences I obtained. Additionally, I attended a highly beneficial workshop on Artificial Intelligence.
Month 9 (July): At the beginning of the month, I completed assignments for the university’s summer semester. I then took some time off for a holiday at home. During this period, I also participated in an online PhD seminar, where I presented my project to professors and fellow PhD students.
Month 10 (August): After returning from holidays, I began working on the new samples collected from Greenland by other group members. Separately, I performed MIC testing on the samples from France that I had previously isolated, sequenced, and identified to assess their antibiotic resistance. Additionally, I enrolled in an online bioinformatics course to expand my knowledge in this area, dedicating a couple of weeks to practicing new techniques. Finally, I prepared for my fieldwork in Austria, which was to be my first major fieldwork experience.
Month 11 (September): During my travel to Austria, I first visited Eurofins, one of Brill Company's industrial partners. This was my first experience visiting an industrial sequencing facility, where I had the opportunity to meet Mashal Alawi, the head of the company. We discussed my project, and he offered valuable suggestions based on his expertise with psychrophilic bacteria. Afterward, I travelled to Austria, where our team conducted fieldwork at the Stubai Glacier. We collected various samples, then processed them by filtering in the laboratory at the University of Innsbruck. Next, I returned to my home lab in Bremen and cultured the filters. At the same time, I designed an experiment to assess the growth of the isolates at different temperatures using OD measurements.
Month 12 (October): I continued working on all current samples, progressing with gene mining on my sequences to identify antibiotic resistance genes. I also organized the data collected from the French samples and compiled it into a presentation file. Additionally, I attended the ICEBIO network event in Copenhagen, where I presented all my data to the attendees.

DC2 LEA FRANCOMME: MAY 2024 - OCToBE 2024
Month 7 (May): I focused on the first sequencing test of Hydrurus foetidus full genome inhouse. As it was Nora’s last month of master thesis, I got the opportunity to develop even more my mentoring skills by checking on the results she produced and proofreading her master thesis.
Month 8 (June): I followed a one-week intensive course on RNA sequencing data analysis at EPFL as well as an intensive course of academic writing over two weeks. Aside from classes, I focused on writing the introduction of the research plan (RP) I will have to defend during the candidacy examination that will occur in November.
Month 9 (July): I pursued the writing of my RP with the detailed plan of each of my PhD chapters. Data obtained from winter 2024 fieldwork campaign were preprocessed to start downstream analysis of Hydrurus foetidus microbial community on solid grounds.
Month 10 (August): August was a short month for me due to vacations; when back to the office, I initiated Hydrurus foetidus microbial community data analysis by preprocessing the data and making sure I also prepared the next fieldwork campaign occurring in October.
Month 11 (September): I performed the data analysis of winter 2024 samples 16S sequencing to obtain a first idea of the microbial community associated to Hydrurus foetidus.
Month 12 (October): I achieved the writing of my RP, gave a talk to student in biochemistry major about ICEBIO and my work during the Day of Orientation and Ecological Transition organized at Collège - Lycée de la Planta in Sion. I was also invited to speak at the Annual Congress of the UPBM (Union des professeurs de Physiologie Biochimie et Microbiologie). Finally, to end nicely this month, I met my fellow PhDs from ICEBIO in Copenhagen for the bi- annual meeting.











